4 Data structure
PyFDAP provides a hierarchical object structure to organize the datasets obtained from FDAP experiments and to facilitate data navigation (Figure 1).
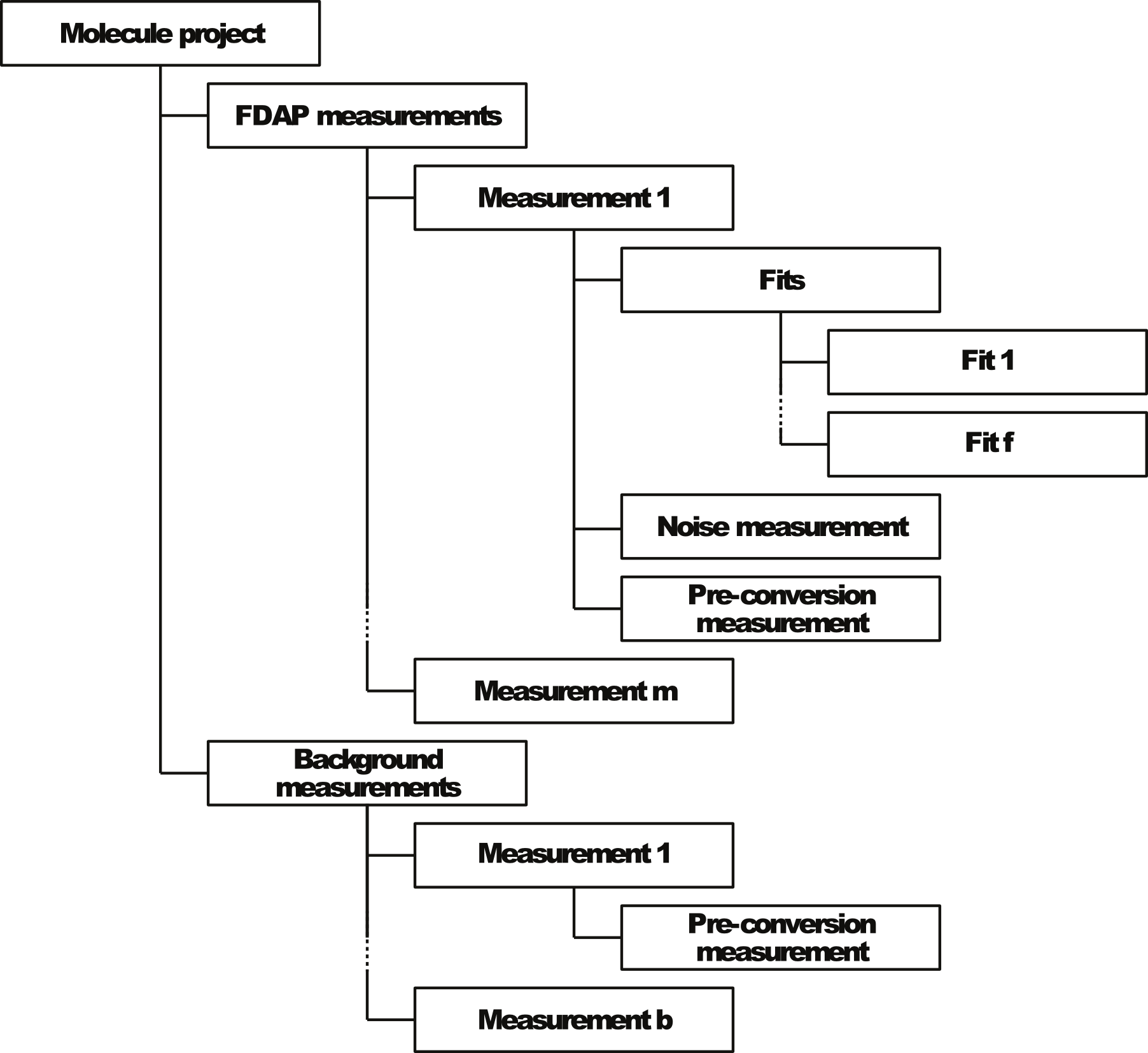
Molecule projects: Replicate experiments with the same protein are grouped into a main molecule
project. PyFDAP can handle multiple molecule projects in one session.
FDAP measurements: Replicate experiments are divided into FDAP and background pre- and
post-conversion measurements. Intra- and extracellular protein stability can be different, and
PyFDAP can import a second dataset that counter-labels intra- or extracellular space. The
separation of fluorescence intensities into intra- or extracellular masks is performed using
the Otsu binarization algorithm (Otsu, 1979). The masks and corresponding datasets
can be investigated inside the PyFDAP GUI by clicking on Data Analysis → Plotting →
Background Dataset. The masks are applied to the images of the photoconverted signal, and the
average intensities in the intra- and extracellular domains and in the entire image are
calculated. Each PyFDAP embryo dataset (FDAP measurement) can have multiple fits
for various regions, using different fitting parameters and different data points to allow
maximum flexibility. The fits are automatically included in the PyFDAP data structure.
Noise measurements: Noise measurements can be imported for each embryo dataset and can be used
to calculate estimates for the baseline of the fit (see Section 6.2).
Pre-conversion measurements: Pre-conversion intensity measurements provide information about the
levels of autofluorescence and can be used to calculate estimates for the baseline of the fit (see
Section 6.2).
Background measurements: Background measurements provide information about the levels of autofluorescence after mock-photoconversion in the presence of unlabeled variants of the protein of interest and can be used to calculate estimates for the baseline of the fit (see Section 6.2).